Handling of Water Molecules in Protein-Ligand Complexes
Waters play an important role in ligand binding and significant gains in affinity and selectivity can be achieved by targeting the right waters.
We have developed WaterRank, a tool with which crystallographic water molecules are assigned a geometric rank score so that researchers can discriminate waters that have a role in binding from waters that can be ignored.
Specifically, water molecules are classified according to their interactions with neighbouring protein atoms and water molecules. For each water, a Rank score is calculated on the basis of the deviation from ideal tetrahedral coordination:
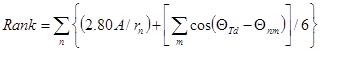
where rn is the distance between the water oxygen and the hydrogen-bonded heavy atom n (n is the number of interacting atoms up to a maximum of 4). This is scaled relative to 2.8 Å, the median hydrogen bonding distance in the Cambridge Structural Database (CSD) for C=O acceptors interacting with OH and NH donors. θTd is the ideal tetrahedral angle (109.5), and θnm is the angle between contact atoms n and m (m = 1 to n – 1).
A maximum of two donors and two acceptors are considered, and any angle less than 60 is rejected from the analysis.
Rank scores can range from 0 (no hydrogen bond) to 6 (four hydrogen bonds in ideal tetrahedral coordination). Water molecules with a Rank score < 2.0, which corresponds to waters not involved in two or more good hydrogen bonds, are ignored. Water-water contacts are included in the calculation of the Rank score if the contacting oxygen atom itself has a score above the threshold of 2.0.
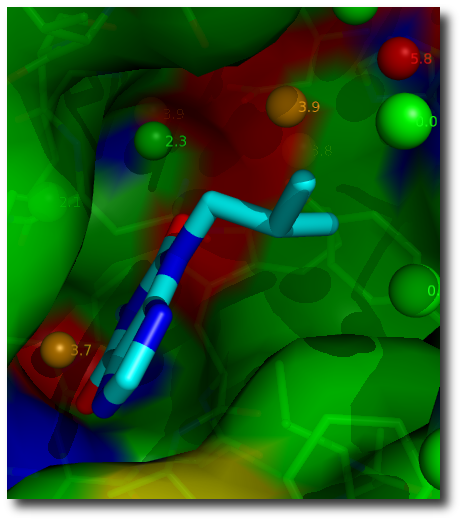
Structural water molecules are color-coded according to the geometric Rank score ranging from green (deemed easily replaceable) to orange (tighter binding).
Color-coding of Rank score:
- green: 0 – 2.3 (easy to replace)
- amber: 2.3 – 4.0 (possible to replace with suitable polar functionalities)
- red: 4.0 – 6.0 (unlikely to replace)
Rank scores provide easy identification of bound water molecules that can be targeted in ligand design experiments
WaterRank is available as a module within ViewConatcts and Viper.